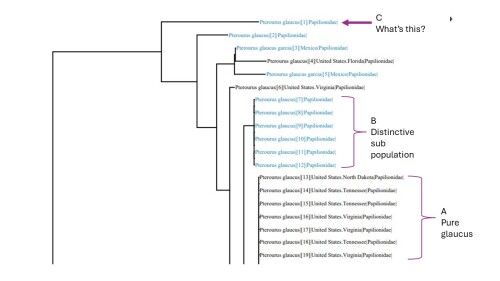
Mitochondrial DNA (MtDNA) is a foundation of current genetic taxonomy. It is not comprehensive, it is not the latest and greatest.
MtDNA, beside being a partial snapshot, has one critical flaw- it is only passed down by the females. None of the male MtDNA is passed down.
So let's look at this.
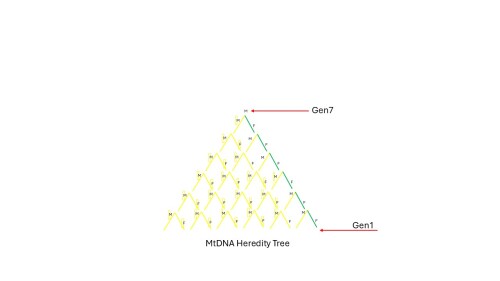
This is a basic heredity tree, showing how each individuals is a product of one male (red) and one female (green) parent. Various types of DNA are passed from each of the parents. You are a sum of your ancestors- sort of.

However, MtDNA is only passed down from the female mother (green.) For the father (red), when the MtDNA from generation X-1 is passed to generation X, that MtDNA "disappears"- it does not get to the generation X individual. NOTE important- but other DNA does, otherwise you'd look like your mother.
Below, all the yellow MtDNA disappears when passed to the next generation because the males do not pass along MtDNA; the green is all that's passed down to the individual marked "Gen 7"
From a more practical perspective, what does this mean? Below, the Gen 1 female passes the MtDNA to the next generation, and IF that individual is a female she too will pass along this MtDNA.
In this example, if the Gen 1 female (and only the female) is Papilio glaucus, then that glaucus MtDNA will continue down the line of females only and the Gen 7 (whether male or female) will have the same MtDNA as the Gen 1 female.
COI is the analysis of some MtDNA, so the heredity of any individual, based on the COI Bar Coding, tracks only mutations in the MtDNA that came along a direct path on the maternal line.
Below, let's say that the green line of females are glaucus; however, the Gen 7 individual also has (obviously) a male father, which has ancestors, and in this case every single one, male and female is Papilio canadensis (yellow.) COI Bar Coding though would concretely put this Gen 7 individual in the Papilio glaucus clade.
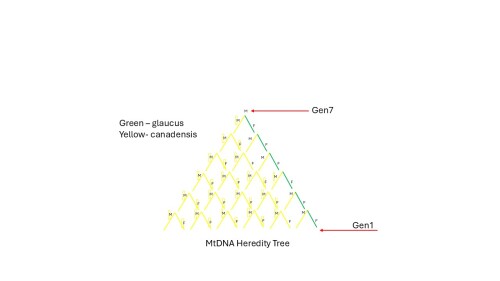
Note too, for the Gen 7 individual, which I've selected to be male, his MtDNA will not be passed down, so all of the MtDNA shown in this tree is at a dead end, so far as his offspring are concerned; it's like they never existed.
So what use is MtDNA? First, it's a pretty good indicator of mutations. Second, it's inexpensive to extract and analyze.
Note too in the above drawings, it's unlikely that two parents produce only one viable offspring, so in any given population the MtDNA will be passed around in that population.
Hope this helps.